Getting started with R Markdown (Optional)
Overview
Teaching: 30 min
Exercises: 15 minQuestions
What is R Markdown?
How can I integrate my R code with text and plots?
How can I convert .Rmd files to .html?
Objectives
Create a .Rmd document containing R code, text, and plots
Create a YAML header to control output
Understand basic syntax of (R)Markdown
Customise code chunks to control formatting
Use code chunks and in-line code to create dynamic, reproducible documents
R Markdown
R Markdown is a flexible type of document that allows you to seamlessly combine executable R code, and its output, with text in a single document. These documents can be readily converted to multiple static and dynamic output formats, including PDF (.pdf), Word (.docx), and HTML (.html).
The benefit of a well-prepared R Markdown document is full reproducibility. This also means that, if you notice a data transcription error, or you are able to add more data to your analysis, you will be able to recompile the report without making any changes in the actual document.
The rmarkdown package comes pre-installed with RStudio, so no action is necessary.
 Image credit: Allison Horst
Image credit: Allison Horst
Creating an R Markdown file
To create a new R Markdown document in RStudio, click File -> New File -> R Markdown:
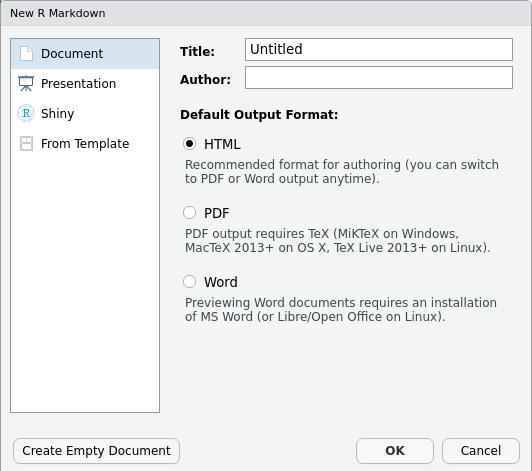
Then click on ‘Create Empty Document’. Normally you could enter the title of your document, your name (Author), and select the type of output, but we will be learning how to start from a blank document.
Basic components of R Markdown
To control the output, a YAML (YAML Ain’t Markup Language) header is needed:
---
title: "My Awesome Report"
author: "Emmet Brickowski"
date: ""
output: html_document
---
The header is defined by the three hyphens at the beginning (---) and the
three hyphens at the end (---).
In the YAML, the only required field is the output:, which specifies the type
of output you want. This can be an html_document, a pdf_document, or a
word_document. We will start with an HTML doument and discuss the other
options later.
The rest of the fields can be deleted, if you don’t need them. After the header,
to begin the body of the document, you start typing after the end of the YAML
header (i.e. after the second ---).
Markdown syntax
Markdown is a popular markup language that allows you to add formatting elements
to text, such as bold, italics, and code. The formatting will not be
immediately visible in a markdown (.md) document, like you would see in a Word
document. Rather, you add Markdown syntax to the text, which can then be
converted to various other files that can translate the Markdown syntax.
Markdown is useful because it is lightweight, flexible, and platform
independent.
Some platforms provide a real time preview of the formatting, like RStudio’s visual markdown editor (available from version 1.4).
First, let’s create a heading! A # in front of text indicates to Markdown that
this text is a heading. Adding more #s make the heading smaller, i.e. one # is
a first level heading, two ##s is a second level heading, etc. upto the 6th level heading.
# Title
## Section
### Sub-section
#### Sub-sub section
##### Sub-sub-sub section
###### Sub-sub-sub-sub section
(only use a level if the one above is also in use)
Since we have already defined our title in the YAML header, we will use a section heading to create an Introduction section.
## Introduction
You can make things bold by surrounding the word with double asterisks,
**bold**, or double underscores, __bold__; and italicize using single
asterisks, *italics*, or single underscores, _italics_.
You can also combine bold and italics to write something really
important with triple-asterisks, ***really***, or underscores, ___really___;
and, if you’re feeling bold (pun intended), you can also use a combination of
asterisks and underscores, **_really_**, **_really_**.
To create code-type font, surround the word with backticks,
`code-type`.
Now that we’ve learned a couple of things, it might be useful to implement them:
## Introduction
This report uses the **tidyverse** package along with the *SAFI* dataset,
which has columns that include:
Then we can create a list for the variables using -, +, or * keys.
## Introduction
This report uses the **tidyverse** package along with the *SAFI* dataset,
which has columns that include:
- village
- interview_date
- no_members
- years_liv
- respondent_wall_type
- rooms
You can also create an ordered list using numbers:
1. village
2. interview_date
3. no_members
4. years_liv
5. respondent_wall_type
6. rooms
And nested items by tab-indenting:
- village
+ Name of village
- interview_date
+ Date of interview
- no_members
+ How many family members lived in a house
- years_liv
+ How many years respondent has lived in village or neighbouring village
- respondent_wall_type
+ Type of wall of house
- rooms
+ Number of rooms in house
For more Markdown syntax see the following reference guide.
Now we can render the document into HTML by clicking the Knit button in the top of the Source pane (top left), or use the keyboard shortcut Ctrl+Shift+K on Windows and Linux, and Cmd+Shift+K on Mac. If you haven’t saved the document yet, you will be prompted to do so when you Knit for the first time.
Writing an R Markdown report
Now we will add some R code from our previous data wrangling and visualisation, which means we need to make sure tidyverse is loaded. It is not enough to load tidyverse from the console, we will need to load it within our R Markdown document. The same applies to our data. To load these, we will need to create a ‘code chunk’ at the top of our document (below the YAML header).
A code chunk can be inserted by clicking Code > Insert Chunk, or by using the keyboard shortcuts Ctrl+Alt+I on Windows and Linux, and Cmd+Option+I on Mac.
The syntax of a code chunk is:
```{r chunk-name}
Here is where you place the R code that you want to run.
```
An R Markdown document knows that this text is not part of the report from the
``` that begins and ends the chunk. It also knows that the code
inside of the chunk is R code from the r inside of the curly braces ({}).
After the r you can add a name for the code chunk . Naming a chunk is
optional, but recommended. Each chunk name must be unique, and only contain
alphanumeric characters and -.
```{r package-data, echo = FALSE, message = FALSE, purl = FALSE} library(tidyverse) interviews <- read_csv(“data/SAFI_clean.csv”, na = “NULL”)
To load **tidyverse** and our `SAFI_clean.csv` file, we will insert a chunk and
call it 'setup'. Since we don't want this code or the output to show in our
knitted HTML document, we add an `include = FALSE` option after the code chunk
name (`{r setup, include = FALSE}`).
<pre>
```{r setup, include = FALSE}
library(tidyverse)
library(here)
interviews <- read_csv(here("data/SAFI_clean.csv"), na = "NULL")
```
</pre>
> ## Important Note!
>
> The file paths you give in a .Rmd document, e.g. to load a .csv file, are
> relative to the .Rmd document, **not** the project root.
>
> As suggested in the Starting with Data episode, we highly recommend the use of
the `here()` function to keep the file paths consistent within your project.
{: .callout}
## Insert table
Next, we will re-create a table from the Data Wrangling episode which
shows the average household size grouped by `village` and `memb_assoc`.
We can do this by creating a new code chunk and calling it 'interview-tbl'.
Or, you can come up with something more creative (just remember to stick to the
naming rules).
It isn't necessary to **Knit** your document every time you want to see the
output. Instead you can run the code chunk with the green triangle in the top
right corner of the the chunk, or with the keyboard shortcuts:
<kbd>Ctrl</kbd>+<kbd>Alt</kbd>+<kbd>C</kbd> on Windows and Linux, or
<kbd>Cmd</kbd>+<kbd>Option</kbd>+<kbd>C</kbd> on Mac.
To make sure the table is formatted nicely in our output document, we will need
to use the `kable()` function from the **knitr** package. The `kable()` function
takes the output of your R code and knits it into a nice looking HTML table. You
can also specify different aspects of the table, e.g. the column names, a
caption, etc.
Run the code chunk to make sure you get the desired output.
```{r interview-tbl, message=FALSE, purl=FALSE}
interviews %>%
filter(!is.na(memb_assoc)) %>%
group_by(village, memb_assoc) %>%
summarize(mean_no_membrs = mean(no_membrs)) %>%
knitr::kable(caption = "We can also add a caption.",
col.names = c("Village", "Member Association",
"Mean Number of Members"))
Customising chunk output
We mentioned using include = FALSE in a code chunk to prevent the code and
output from printing in the knitted document. There are additional options
available to customise how the code-chunks are presented in the output document.
The options are entered in the code chunk after chunk-nameand separated by
commas, e.g. {r chunk-name, eval = FALSE, echo = TRUE}.
| Option | Options | Output |
|---|---|---|
eval |
TRUE or FALSE |
Whether or not the code within the code chunk should be run. |
echo |
TRUE or FALSE |
Choose if you want to show your code chunk in the output document. echo = TRUE will show the code chunk. |
include |
TRUE or FALSE |
Choose if the output of a code chunk should be included in the document. FALSE means that your code will run, but will not show up in the document. |
warning |
TRUE or FALSE |
Whether or not you want your output document to display potential warning messages produced by your code. |
message |
TRUE or FALSE |
Whether or not you want your output document to display potential messages produced by your code. |
fig.align |
default, left, right, center |
Where the figure from your R code chunk should be output on the page |
Tip
- The default settings for the above chunk options are all
TRUE.- The default settings can be modified per chunk, or with
knitr::opts_chunk$set(),- Entering
knitr::opts_chunk$set(echo = FALSE)will change the default of value ofechotoFALSEfor every code chunk in the document.
Exercise
Play around with the different options in the chunk with the code for the table, and re-Knit to see what each option does to the output.
What happens if you use
eval = FALSEandecho = FALSE? What is the difference between this andinclude = FALSE?Solution to Exercise
Create a chunk with
{r eval = FALSE, echo = FALSE}, then create another chunk with{r include = FALSE}to compare.eval = FALSEandecho = FALSEwill neither run the code in the chunk, nor show the code in the knitted document. The code chunk essentially doesn’t exist in the knitted document as it was never run. Whereasinclude = FALSEwill run the code and store the output for later use.
In-line R code
Now we will use some in-line R code to present some descriptive statistics.
To use in-line R-code, we use the same backticks that we used in the Markdown
section, with an r to specify that we are generating R-code. The difference
between in-line code and a code chunk is the number of backticks. In-line R code
uses one backtick (`r`), whereas code chunks use three backticks
(``` r ```).
For example, today’s date is `r Sys.Date()`, will be rendered as:
today’s date is r Sys.Date().
The code will display today’s date in the output document (well, technically the
date the document was last knitted).
The best way to use in-line R code, is to minimise the amount of code you need to produce the in-line output by preparing the output in code chunks. Let’s say we’re interested in presenting the average household size in a village.
```{r in-line-code, purl=FALSE}
create a summary data frame with the mean household size by village
mean_household <- interviews %>% group_by(village) %>% summarize(mean_no_membrs = mean(no_membrs))
and select the village we want to use
mean_chirodzo <- mean_household %>% filter(village == “Chirodzo”)
Now we can make an informative statement on the means of each village, and include
the mean values as in-line R-code. For example:
The average household size in the village of Chirodzo is
`r round(mean_chirodzo$mean_no_members, 2)`
becomes...
The average household size in the village of Chirodzo is
`r round(mean_chirodzo$mean_no_membrs, 2)`.
Because we are using in-line R code instead of the actual values, we have created
a dynamic document that will automatically update if we make changes to the dataset
and/or code chunks.
## Plots
Finally, we will also include a plot, so our document is a little more colourful
and a little less boring. We will use the `interview_plotting` data from the
previous episode.
If you were unable to complete the previous lesson or did not save the data,
then you can create it in a new code chunk.
```{r interview-plotting-data, purl = FALSE, eval = TRUE}
## Not run, but can be used to load in data from previous lesson!
interviews_plotting <- interviews %>%
## pivot wider by items_owned
separate_rows(items_owned, sep = ";") %>%
## if there were no items listed, changing NA to no_listed_items
replace_na(list(items_owned = "no_listed_items")) %>%
mutate(items_owned_logical = TRUE) %>%
pivot_wider(names_from = items_owned,
values_from = items_owned_logical,
values_fill = list(items_owned_logical = FALSE)) %>%
## pivot wider by months_lack_food
separate_rows(months_lack_food, sep = ";") %>%
mutate(months_lack_food_logical = TRUE) %>%
pivot_wider(names_from = months_lack_food,
values_from = months_lack_food_logical,
values_fill = list(months_lack_food_logical = FALSE)) %>%
## add some summary columns
mutate(number_months_lack_food = rowSums(select(., Jan:May))) %>%
mutate(number_items = rowSums(select(., bicycle:car)))
Exercise
Create a new code chunk for the plot, and copy the code from any of the plots we created in the previous episode to produce a plot in the chunk. I recommend one of the colourful plots.
If you are feeling adventurous, you can also create a new plot with the
interviews_plottingdata frame.Solution to Exercise
{r my-fancy-plot, answer = TRUE, purl = FALSE} interviews_plotting %>% ggplot(aes(x = respondent_wall_type)) + geom_bar(aes(fill = village))
We can also create a caption with the chunk option fig.cap.
```{r chunk-name, fig.cap = "I made this plot while attending an
awesome Data Carpentries workshop where I learned a ton of cool stuff!"}
Code for plot
```
…or, ideally, something more informative.
```{r caption-plot, eval = TRUE, fig.cap = “I made this plot while attending an awesome Data Carpentries workshop where I learned a ton of cool stuff!”} interviews_plotting %>% ggplot(aes(x = respondent_wall_type)) + geom_bar(aes(fill = village), position = “dodge”) + labs(x = “Type of Wall in Home”, y = “Count”, fill = “Village Name”) + scale_fill_viridis_d() # add colour deficient friendly palette
## Other output options
You can convert R Markdown to a PDF or a Word document (among others).
Click the little triangle next to the **Knit** button to get a drop-down menu.
Or you could put `pdf_document` or `word_document` in the initial header of the file.
title: “My Awesome Report” author: “Emmet Brickowski” date: “” output: word_document — ```
Note: Creating PDF documents
Creating .pdf documents may require installation of some extra software. The R package
tinytexprovides some tools to help make this process easier for R users. Withtinytexinstalled, runtinytex::install_tinytex()to install the required software (you’ll only need to do this once) and then when you Knit to pdftinytexwill automatically detect and install any additional LaTeX packages that are needed to produce the pdf document. Visit the tinytex website for more information.
Resources
- Knitr in a knutshell tutorial
- Dynamic Documents with R and knitr (book)
- R Markdown documentation
- R Markdown cheat sheet
- Getting started with R Markdown
- Markdown tutorial
- R Markdown: The Definitive Guide (book by Rstudio team)
- Reproducible Reporting
- The Ecosystem of R Markdown
- Introducing Bookdown
Key Points
R Markdown is a useful language for creating reproducible documents combining text and executable R-code.
Specify chunk options to control formatting of the output document